ParPyDTK2 Python API¶
Default¶
Main module interface of ParPyDTK2
-
parpydtk2.B2G¶ bool – boolean flag of
Truedenotes transferring direction frombluetogreen
-
parpydtk2.G2B¶ bool – boolean flag of
Falsedenotes transferring direction fromgreentoblue
-
parpydtk2.MMLS¶ int – flag (0) represents using modified moving least square method
-
parpydtk2.SPLINE¶ int – flag (1) represents using spline interpolation method
-
parpydtk2.N2N¶ int – flag (2) represents using nearest node projection method
-
parpydtk2.AWLS¶ int – flag (3) represents using adaptive weighted least square method
-
parpydtk2.N2N_MATCH¶ int – flag (4) represents using matching n2n method
-
parpydtk2.WENDLAND2¶ int – flag (0) represents using Wendland 2nd-order RBF weights
-
parpydtk2.WENDLAND4¶ int – flag (1) represents using Wendland 4th-order RBF weights
-
parpydtk2.WENDLAND6¶ int – flag (2) represents using Wendland 6th-order RBF weights
-
parpydtk2.WU2¶ int – flag (3) represents using Wu 2nd-order RBF weights
-
parpydtk2.WU4¶ int – flag (4) represents using Wu 4th-order RBF weights
-
parpydtk2.WU6¶ int – flag (5) represents using Wu 6th-order RBF weights
-
parpydtk2.BUHMANN3¶ int – flag (6) represents using Buhmann 3rd-order RBF weights
-
parpydtk2.WENDLAND21¶ int – flag (7) represents using Wendland 2nd-order 1st dimension RBF weights
-
parpydtk2.create_imeshdb_pair(comm=None)[source]¶ Create a pair of interface mesh databases
This is a convenient and safe way to create a pair of imeshdbs, i.e.
blueandgreenparticipants with a unified communicator. Please use this API instead of directly creatingIMeshDB.Parameters: comm (MPI.Comm (optional)) – MPI communicator, default is NoneorMPI_COMM_WORLD.Returns: blueandgreenparticipantsReturn type: tuple of IMeshDBExamples
Create mesh databases with
MPI_COMM_WORLD>>> from parpydtk2 import * >>> blue, green = create_imeshdb_pair()
Create mesh databases with explicit communicator
>>> from mpi4py import * >>> from parpydtk2 import * >>> comm = MPI.COMM_WORLD >>> blue, green = create_imeshdb_pair(comm)
Error Handling¶
The error handler module
-
parpydtk2.error_handle.ERROR_CODE¶ int – set this to nonzero values one exceptions have been raised
Examples
>>> import parpydtk2 as dtk
>>> try:
... # your programs here
... except Expection:
... dtk.error.ERROR_CODE = 1
... raise
Interface Mesh & Mapper¶
-
class
parpydtk2.IMeshDB(comm=None)¶ Interface mesh database
ParPyDTK2 utilizes MOAB as the underlying mesh database. MOAB is an array based mesh library that is adapted by DTK2. With array based mesh library, the memory usage and computational cost are lower than typical pointer based data structure. The mesh concept in this work is simple since only meshless methods are ultilized, the only additional attribute one needs is the global IDs/handles, which are used by both MOAB and DTK2. For most applications, the global IDs can be computed offline.
One thing is not directly supported by IMeshDB is I/O. However, since this is a Python module and only points clouds are needed, one can easily uses a tool (e.g. meshio) to load the mesh.
-
comm¶ MPI.Comm – MPI communicator
-
ranks¶ int – size of comm
-
rank¶ int – rank of comm
-
size¶ int – point cloud size, i.e. number of vertices
-
gsize¶ int – global point cloud size
-
bbox¶ np.ndarray – local bounding box array of shape (2,3)
-
gbbox¶ np.ndarray – global bounding box array of shape (2,3)
Constructor
Parameters: comm (MPI.Comm (optional)) – if no communicator or Noneis passed in, thenMPI_COMM_WORLDwill be usedExamples
>>> # implicit communciator >>> import parpydtk2 as dtk >>> mdb = dtk.IMeshDB()
>>> # explicit communicator >>> from mpi4py import MPI >>> import parpydtk2 as dtk >>> mdb = dtk.IMeshDB(MPI.COMM_WOLRD)
Notes
Since the mesh database participants appear at least in pairs, a prefered way to construct IMeshDB is to use the wrapper API
create_imeshdb_pair(), e.g.>>> from parpydtk2 import * >>> blue, green = create_imeshdb_pair()
-
assign_field(self, unicode field_name, ndarray values)¶ Assign values to a field
Note
values size must be at least size*dim
Parameters: - field_name (str) – name of the field
- values (np.ndarray) – input source values
See also
extract_field()- extract value from a field
size- check the size of a mesh set
-
assign_gids(self, __Pyx_memviewslice gids)¶ Assign global IDs
Internally, both DTK and MOAB use so-called global IDs/handles communications. Each node has its own local IDs/handles and a unique global ID.
Parameters: gids (np.ndarray) – global IDs See also
create_vertices()- create vertices
extract_gids()- extract global IDs
-
bbox np.ndarray – local bounding box
The bounding box is stored simply in a 2x3 array, where the first row stores the maximum bounds while minimum bounds for the second row.
Warning
Bounding box is valid only after
finish_create().See also
gbbox- global bounding box
-
begin_create(self)¶ Begin to create/manupilate the mesh
This function must be called in order to let the mesh databse be aware that you will create meshes.
See also
finish_create()- finish creating mesh
-
comm MPI.Comm – communicator
-
create_field(self, unicode field_name, int dim=1)¶ Create a data field for solution transfer
This is the core function to register a field so that you can then transfer its values to other domains. The
dimparameter determines the data type of the field. By default, it’s 1, i.e. scalar fields. For each node, a tensor of (1x``dim``) can be registered. For instance, to transfer forces and displacements in FSI applications,dimis 3 (for 3D problems).Parameters: - field_name (str) – name of the field
- dim (int) – dimension of the field, i.e. scalar, vector, tensor
Examples
>>> from parpydtk2 import * >>> mdb1 = IMeshDB() >>> mdb1.begin_create() >>> # creating the meshdb >>> mdb1.finish_create() >>> mdb1.create_field('heat flux')
-
create_vertices(self, __Pyx_memviewslice coords)¶ Create a set of coordinates
Note
The
coordsmust be C-ordering with ndim=2!Parameters: coords (np.ndarray) – nx3 coordinates in double precision See also
assign_gids()- assign global IDs
extract_vertices()- extract vertex coordinates
Examples
>>> from parpydtk2 import * >>> import numpy as np >>> mdb1 = IMeshDB() >>> mdb1.begin_create() >>> verts = np.zeros((2,3)) # two nodes >>> verts[1][0] = 1.0 >>> mdb1.create_vertices(verts)
-
created(self)¶ Check if the mesh database has been created or not
Returns: Trueiffinish_create()has been calledReturn type: bool
-
empty(self)¶ Check if this is an empty partition
-
extract_field(self, unicode field_name, __Pyx_memviewslice buffer=None, reshape=False)¶ Extact the values from a field
Warning
if buffer is passed in, it must be 1D
Parameters: - field_name (str) – name of the field
- buffer (np.ndarray) – 1D buffer
- reshape (bool) – True if we reshape the output, only for vectors/tensors
Returns: field data values
Return type: np.ndarray
-
extract_gids(self)¶ Extract global IDs/handles
Warning
This function should be called once you have finished
assign_gids().Returns: array of sizethat stores the integer IDsReturn type: np.ndarray
-
extract_vertices(self)¶ Extract coordinate
Warning
This function should be called once you have finished
create_vertices().Returns: (nx3) array that stores the coordinate values Return type: np.ndarray See also
size- get the mesh size
-
field_dim(self, unicode field_name)¶ Check the field data dimension
Parameters: field_name (str) – name of the field Returns: data field dimension of field_name Return type: int
-
finish_create(self, trivial_gid=True)¶ finish mesh creation
This method finalizes the interface mesh database by communicating the bounding boxes and empty partitions. Also, setting up the DTK managers happens here.
Warning
You must call this function once you have done with manupilating the mesh, i.e. vertices and global IDs.
Parameters: trivial_gid (bool) – True if we use MOAB trivial global ID computation Notes
By
trivial_gid, it means simply assigning the global IDs based on the size of the mesh. This is useful in serial settings or transferring solutions from a serial solver to a partitioned one.
-
gbbox np.ndarray – global bounding box
The bounding box is stored simply in a 2x3 array, where the first row stores the maximum bounds while minimum bounds for the second row.
Warning
Bounding box is valid only after
finish_create().See also
bbox- local bounding box
-
gsize int – Get the global point cloud size
-
has_empty(self)¶ Check if an empty partition exists
-
has_field(self, unicode field_name)¶ Check if a field exists
Parameters: field_name (str) – name of the field Returns: True if this meshdb has field_name Return type: bool
-
rank int – get the rank
-
ranks int – Get the communicator size
-
resolve_empty_partitions(self, unicode field_name)¶ Resolve asynchronous values on empty partitions
ParPyDTK2 doesn’t expect
assign_field()should be called collectively. Therefore, a collective call must be made for resolving empty partitions.Warning
This must be called collectively even on empty partitions
Note
You should call this function following assignment
Parameters: field_name (str) – name of the field Examples
>>> if rank == 0: ... mdb.assign_field('flux', values) >>> mdb.resolve_empty_partitions('flux')
Notes
This API is not available in C++ level, therefore, one needs to implement this if he/she wants to use the C++ API.
-
size int – Get the size of a set
-
-
class
parpydtk2.Mapper(blue, green, profiling=True, verbose=True, **kwargs)¶ DTK2 wrapper
The meshless methods in DTK2, including modified moving least square, spline interpolation and nearest node projection methods are wrapped within this class.
In addition, if you build DTK2 from UNIFEM/CHIAO45 forked verions, then you can use the adaptive weighted least square fitting method.
-
comm¶ MPI.Comm – MPI communicator
-
ranks¶ int – size of comm
-
rank¶ int – rank of comm
-
dimension¶ int – spatial dimension
-
method¶ int – method flag, either MMLS, SPLINE, or N2N
-
basis¶ int – flag of basis function for weighting schemes used by MMLS and SPLINE
-
radius_b¶ float – radius used for searching on blue_mesh
-
radius_g¶ float – radius used for searching on green_mesh
-
leaf_b¶ int – kd-tree leaf size of blue_mesh
-
leaf_g¶ int – kd-tree leaf size of green_mesh
-
rho¶ double – local Vandermonde system row scaling factor
Constructor
Parameters: - blue_mesh (
IMeshDB) – blue mesh participant - green_mesh (
IMeshDB) – green mesh participant - profiling (bool (optional)) – whether or not do timing report, default is
True. - verbose (bool (optional)) – whether or not verbose printing, default is
True. - stat_file (str (optional)) – file for storing profiling information
Examples
>>> from parpydtk2 import * >>> blue, green = IMeshDB(), IMeshDB() >>> # initialize blue and green >>> mapper = Mapper(blue=blue,green=green) >>> # do work with mapper
-
awls_conf(self, **kwargs)¶ Configuration of Adaptive Weighted Least Square Fitting
Parameters: - basis (int (optional)) – basis weighting scheme, default is WENDLAND21
- rho (float (optional)) – number of rows in the local Vandermonde system, i.e. rho*col
- ref_r_b (float (optional)) – reference user-specified blue radius, i.e.
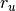 for blue
for blue - ref_r_g (float (optional)) – reference user-specified green radius, i.e.
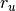 for green
for green - dim (int (optional)) – topological dimension, default is the surface dimension
- verbose (bool (optional)) – print verbose information/warning messages, default is
False - alpha (float) – the
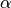 parameter
parameter - beta (float) – the
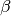 parameter
parameter - _ind_file (str (optional)) – indicator result file, used in chiao45 dtk
Notes
_ind_fileis for internal use to fine tune the parametersigma. It will not be enabled in release build.
-
basis int – Get the basis function flag
See also
method- get the method tag
-
begin_initialization(self, **kwargs)¶ Initialization starter
This is a must-call function in order to indicate mapper that you are about to initialize/register coupling fields
See also
register_coupling_fields()- register coupled fields
end_initialization()- finish initialization
-
begin_transfer(self, **kwargs)¶ Transfer starter
This is a must-call function to inidate the beginning of a transferring block
See also
end_transfer()- transfer closer
tranfer_data()- transfer a coupled data fields
-
blue_mesh IMeshDB– blue mesh
-
comm MPI.Comm – Get the communicator
-
dimension int – Get the problem dimension
Note
this is the spacial dimension
-
enable_mmls_auto_conf(self, *, ref_r_b=None, ref_r_g=None, dim=None, verbose=False, **kwargs)¶ Automatically set up radius parameter for MMLS
Warning
This method should be used only when the underlying DTK2 installation is from CHIAO45 forked version. Otherwise, you should always manully configure the radius parameters.
The following strategy is performed:
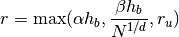
where
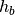 is the the the maximum edge length of the global
bounding box (
is the the the maximum edge length of the global
bounding box (gbbox);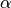 is
some ratio, say 0.1 (10%);
is
some ratio, say 0.1 (10%); 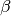 is the scaling factor the
the estimated mesh size, which is given by
is the scaling factor the
the estimated mesh size, which is given by
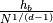 and
and 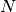 is the global mesh size
(
is the global mesh size
(gsize);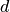 is the spatial
dimension; the last parameter
is the spatial
dimension; the last parameter 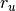 is provided by the user thus
optional.
is provided by the user thus
optional.For UNIFEM/CHIAO45 DTK2, this parameter should be relatively large, because the final points in the Vandermonde system is determined by the column size, so that larger radius means that the system has larger candidate pool.
Warning
Regarding the spatial dimension, since this packages is mainly for interface/surface coupling thus the actual dimension is assumed to be one less than the spatial dimension, i.e. surface topological dimension. If this is not the case, the user needs to explicit pass in the dimension to override this default behavior.
Parameters: - ref_r_b (float (optional)) – reference user-specified blue radius, i.e.
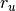 for blue
for blue - ref_r_g (float (optional)) – reference user-specified green radius, i.e.
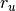 for green
for green - dim (int (optional)) – topological dimension, default is the surface dimension
- verbose (bool (optional)) – print verbose information/warning messages, default is
False - alpha (float) – the
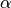 parameter
parameter - beta (float) – the
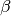 parameter
parameter
See also
is_dtk2_backend()- check backend installation of DTK2
- ref_r_b (float (optional)) – reference user-specified blue radius, i.e.
-
end_initialization(self, **kwargs)¶ Initialization closer
This is a must-call function in order to tell the mapper we are ready
See also
begin_initialization()- initialization starter
-
end_transfer(self, **kwargs)¶ Transfer closer
This is a must-call function to indicate we have finished a sequence of transferring requests
See also
begin_transfer()- transfer starter
-
green_mesh IMeshDB– green mesh
-
has_coupling_fields(self, unicode bf, unicode gf, bool direct)¶ Check if a coupled fields exists
Returns: Trueif (bf,gf) exists in thedirectdirectionReturn type: bool
-
static
is_dtk2_backend()¶ Check if the underlying DTK2 library is chiao45 forked version
Returns: Falseif the user’s DTK2 is from chiao45 forked repoReturn type: bool
-
leaf_b int – get the leaf size of blue mesh for kd-tree
Warning
This attribute only works when the underlying DTK2 is installed from UNIFEM or CHIAO45 forked versions.
See also
leaf_g- green leaf size of kd-tree
-
leaf_g int – get the leaf size of the green mesh for kd-tree
Warning
This attribute only works when the underlying DTK2 is installed from CHIAO45 forked versions.
See also
leaf_b- blue leaf size of kd-tree
-
method int – Get the method tag
See also
basis- the basis function and order attribute
-
static
parameter_keys()¶
-
radius_b float – physical domain radius support for blue mesh
Note
if blue does not use RBF-search, then -1.0 returned
See also
radius_g- green radius
-
radius_g float – physical domain radius support for green mesh
Note
if green does not use RBF-search, then -1.0 returned
See also
radius_b- blue radius
-
rank int – Check “my” rank
-
ranks int – Check the total process number
See also
rank- “my” rank
-
register_coupling_fields(self, unicode bf, unicode gf, bool direct, **kwargs)¶ register a coupled fields
Note
we use boolean to indicate direction
Parameters: - bf (str) – blue mesh field name
- gf (str) – green mesh field name
- direct (bool) – True for blue->green, False for the opposite
See also
transfer_data()- transfer a coupled data fields
-
rho float – local scaling factor
-
set_matching_flag_n2n(self, bool matching)¶ Set the matching flag for N2N
Note
this function will not throw even if you dont use n2n
Parameters: matching (bool) – True if the interfaces are matching
-
transfer_data(self, unicode bf, unicode gf, bool direct, **kwargs)¶ Transfer (bf, gf) in the
directdirectionNote
Parameters
resolve_discandsigmaonly work with UNIFEM/CHIAO45 DTK and AWLS method.Parameters: - bf (str) – blue mesh field name
- gf (str) – green mesh field name
- direct (bool) –
Truefor blue->green,Falsefor the opposite - resolve_disc (bool (optional)) –
Trueif do post-processing for resolving non-smooth functions - sigma (float (optional)) – threshold used in smoothness indicator
See also
register_coupling_fields()- register coupled fields
-